
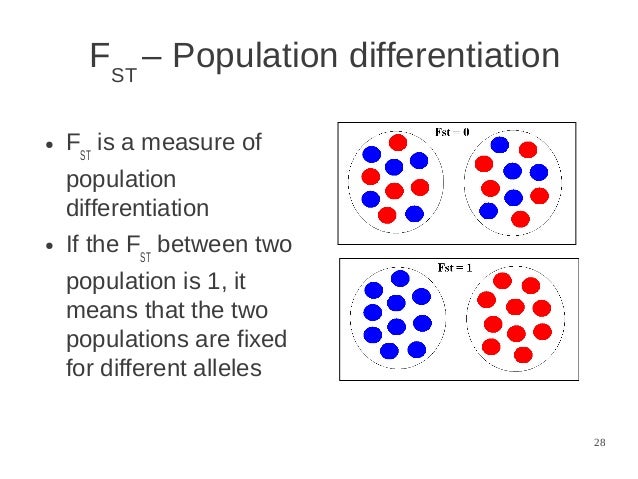
Table 4.2 Frequencies of alternative wing genotypes in two hypothetical subpopulations of the scarlet tiger moth Since FST measures the extent to which populations have differentiated from one another, this is the F-statistic with which we are most concerned in this chapter (see Box 4.2). The relationship between the three statistics is given as: Where Ht and HI are the same as in Equations 4.4 and 4.5. Fit is therefore influenced by both non-random mating within a subpopulation (FIS) and population subdivision (FST), and is calculated as: This provides an overall inbreeding coefficient for an individual by measuring the heterozygosity of an individual relative to the total population. The third F-statistic, which is used much less frequently than the other two, is FIT. Where HS is the same as in Equation 4.4 and Ht is the expected heterozygosity of the total population. it is a measure of the degree of inbreeding within a subpopulation relative to the total population (total population here meaning all of the subpopulations combined), and reflects the probability that two alleles drawn at random from within a subpopulation are identical by descent. The second F-statistic is FST (also known as the fixation index), and this provides an estimate of the genetic differentiation between subpopulations. Where HI is the observed heterozygosity in a subpopulation at the time of investigation (individual heterozygosity) and HS is the heterozygosity that would be expected if the subpopulation was in HWE (subpopulation heterozygosity) This reflects the probability that two alleles within the same individual are identical by descent, and is the same as the inbreeding coefficient F that was introduced in Chapter 3. The first F-statistic, FIS, measures the degree of inbreeding within individuals relative to the rest of their subpopulation. Note that in describing these levels we will follow the convention set by the literature underlying the theory of F-statistics, and refer to spatially discrete samples as subpopulations instead of populations elsewhere we will revert to the more common practice of referring to discrete breeding units as populations. F-Statistics use inbreeding coefficients to describe the partitioning of genetic variation within and among populations and can be calculated at three different levels. Perhaps the most common method for quantifying the genetic differentiation between populations is based on F-statistics, which were developed by Wright (1951).


 0 kommentar(er)
0 kommentar(er)
